Triplet STDP synapse tutorial
In this tutorial, we will learn to formulate triplet rule (which considers sets of three spikes, i.e., two presynaptic and one postsynaptic spikes or two postsynaptic and one presynaptic spikes) for Spike Timing-Dependent Plasticity (STDP) learning model using NESTML and simulate it with NEST simulator.
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import nest
import numpy as np
import os
import re
from pynestml.frontend.pynestml_frontend import to_nest, install_nest
np.set_printoptions(suppress=True)
NEST_INSTALL_PATH = nest.ll_api.sli_func("statusdict/prefix ::")
Early experiments in Bi and Poo (1998) [1] have shown that a sequence of \(n\) pairs of “pre then post” spikes result in synaptic potentiation and \(n\) pairs of “post then pre” result in synaptic depression. Later experiments have shown that these pairs of spikes do not necessarily describe the synaptic plasiticity behavior. Other variables like calcium concentration or postsynaptic membrane potential play an important role in potentiation or depression.
Experiments conducted by Wang et al., [2] and Sjöström et al., [3] using triplet and quadruplets of spikes show that the classical spike-dependent synaptic plasticity alone cannot explain the results of the experiments. The triplet STDP model formulated by Pfister and Gerstner in [4] assume that a combination of pairs and triplets of spikes triggers the synaptic plasticity and thus reproduce the experimental results.
Triplet STDP model
The synaptic transmission is formulated by a set of four differential equations as defined below.
Here, \(r_1\) and \(r_2\) are the trace variables that detect the presynaptic events when a spike occurs at the presynaptic neuron at time \(t^{pre}\). These variables increase when a presynaptic spike occurs and decrease back to 0 otherwise with time constants \(\tau_+\) and \(\tau_x\) respectively. Similarly, the variables \(o_1\) and \(o_2\) denote the trace variables that detect the postsynaptic events at a spike time \(t^{post}\). In the absence of a postsynaptic spike, the vairables decrease their value with a time constant \(\tau_-\) and \(\tau_y\) respectively. Presynaptic variables, for example, can be the amount of glutamate bound and the postsynaptic receptors can determine the influx of calcium concentration through voltage-gated \(Ca^{2+}\) channels.
The weight change in the model is formulated as a function of the four trace variables defined above. The weight of the synapse decreases after the presynaptic spikes arrives at \(t^{pre}\) by an amount proportional to the postsynaptic variable \(o_1\) but also depends on the second presynaptic variable \(r_2\).
Similarly, a postsynaptic spike at time \(t^{post}\) increases the weight of the synapse and is dependent on the presynaptic variable \(r_1\) and the postsynaptic variable \(o_2\).
Here, \(A_2^+\) and \(A_2^-\) denote the amplitude of the weight change whenever there is a pre-post pair or a post-pre pair. Similarly, \(A_3^+\) and \(A_3^-\) denote the amplitude of the triplet term for potentiation and depression, respectively.
Generating code with NESTML
Triplet STDP model in NESTML
In this tutorial, we will use a very simple integrate-and-fire model, where arriving spikes cause an instantaneous increment of the membrane potential. Let’s download the model from the NESTML repository so it becomes available locally:
[2]:
import urllib.request
if not os.path.isdir("models"):
os.makedirs("models")
if not os.path.isdir("models/neurons"):
os.makedirs("models/neurons")
if not os.path.isdir("models/synapses"):
os.makedirs("models/synapses")
urllib.request.urlretrieve("https://raw.githubusercontent.com/nest/nestml/master/models/neurons/iaf_psc_delta.nestml",
"models/neurons/iaf_psc_delta.nestml")
[2]:
('models/neurons/iaf_psc_delta.nestml', <http.client.HTTPMessage at 0x7fec6c89df70>)
We will be formulating two types of interactions between the spikes, namely All-to-All interaction and Nearest-spike interaction.
\(\textit{All-to-All Interation}\) - Each postsynaptic spike interacts with all previous postsynaptic spikes and vice versa. All the internal variables \(r_1, r_2, o_1,\) and \(o_2\) accumulate over several postsynaptic spike timimgs.
[3]:
nestml_triplet_stdp_model = '''
synapse stdp_triplet:
state:
w nS = 1 nS
tr_r1 real = 0.
tr_r2 real = 0.
tr_o1 real = 0.
tr_o2 real = 0.
end
parameters:
the_delay ms = 1 ms @nest::delay # !!! cannot have a variable called "delay"
tau_plus ms = 16.8 ms # time constant for tr_r1
tau_x ms = 101 ms # time constant for tr_r2
tau_minus ms = 33.7 ms # time constant for tr_o1
tau_y ms = 125 ms # time constant for tr_o2
A2_plus real = 7.5e-10
A3_plus real = 9.3e-3
A2_minus real = 7e-3
A3_minus real = 2.3e-4
Wmax nS = 100 nS
Wmin nS = 0 nS
end
equations:
tr_r1' = -tr_r1 / tau_plus
tr_r2' = -tr_r2 / tau_x
tr_o1' = -tr_o1 / tau_minus
tr_o2' = -tr_o2 / tau_y
end
input:
pre_spikes nS <- spike
post_spikes nS <- spike
end
output: spike
onReceive(post_spikes):
# increment post trace values
tr_o1 += 1
tr_o2 += 1
# potentiate synapse
w_ nS = w + tr_r1 * ( A2_plus + A3_plus * tr_o2 )
w = min(Wmax, w_)
end
onReceive(pre_spikes):
# increment pre trace values
tr_r1 += 1
tr_r2 += 1
# depress synapse
w_ nS = w - tr_o1 * ( A2_minus + A3_minus * tr_r2 )
w = max(Wmin, w_)
# deliver spike to postsynaptic partner
deliver_spike(w, the_delay)
end
end
'''
[4]:
n_modules_generated = 0
def generate_code_for(nestml_synapse_model: str):
"""Generate code for a given synapse model, passed as a string, in combination with
the iaf_psc_delta model.
NEST cannot yet reload modules. Workaround using counter to generate unique names."""
global n_modules_generated
# append digit to the neuron model name and neuron model filename
with open("models/neurons/iaf_psc_delta.nestml", "r") as nestml_model_file_orig:
nestml_neuron_model = nestml_model_file_orig.read()
nestml_neuron_model = re.sub("neuron\ [^:\s]*:",
"neuron iaf_psc_delta" + str(n_modules_generated) + ":", nestml_neuron_model)
with open("models/neurons/iaf_psc_delta" + str(n_modules_generated) + ".nestml", "w") as nestml_model_file_mod:
print(nestml_neuron_model, file=nestml_model_file_mod)
# append digit to the synapse model name and synapse model filename
nestml_synapse_model_name = re.findall("synapse\ [^:\s]*:", nestml_synapse_model)[0][8:-1]
nestml_synapse_model = re.sub("synapse\ [^:\s]*:",
"synapse " + nestml_synapse_model_name + str(n_modules_generated) + ":", nestml_synapse_model)
with open("models/synapses/" + nestml_synapse_model_name + str(n_modules_generated) + ".nestml", "w") as nestml_model_file:
print(nestml_synapse_model, file=nestml_model_file)
# generate the code for neuron and synapse (co-generated)
module_name = "nestml_" + str(n_modules_generated) + "_module"
to_nest(input_path=["models/neurons/iaf_psc_delta" + str(n_modules_generated) + ".nestml",
"models/synapses/" + nestml_synapse_model_name + str(n_modules_generated) + ".nestml"],
target_path="/tmp/nestml_module",
logging_level="ERROR",
module_name=module_name,
suffix="_nestml",
codegen_opts={"neuron_parent_class": "StructuralPlasticityNode",
"neuron_parent_class_include": "structural_plasticity_node.h",
"neuron_synapse_pairs": [{"neuron": "iaf_psc_delta" + str(n_modules_generated),
"synapse": nestml_synapse_model_name + str(n_modules_generated),
"post_ports": ["post_spikes"]}]})
install_nest("/tmp/nestml_module", NEST_INSTALL_PATH)
# load module into NEST
nest.ResetKernel()
nest.Install(module_name)
mangled_neuron_name = "iaf_psc_delta" + str(n_modules_generated) + "_nestml__with_" + nestml_synapse_model_name + str(n_modules_generated) + "_nestml"
mangled_synapse_name = nestml_synapse_model_name + str(n_modules_generated) + "_nestml__with_iaf_psc_delta" + str(n_modules_generated) + "_nestml"
n_modules_generated += 1
return mangled_neuron_name, mangled_synapse_name, nestml_synapse_model_name
[5]:
# Generate code for All-to-All spike interaction
neuron_model_name, synapse_model_name, nestml_synapse_model_name = \
generate_code_for(nestml_triplet_stdp_model)
[1,GLOBAL, INFO]: List of files that will be processed:
[2,GLOBAL, INFO]: /home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/neurons/iaf_psc_delta0.nestml
[3,GLOBAL, INFO]: /home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/synapses/stdp_triplet0.nestml
[4,GLOBAL, INFO]: Start processing '/home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/neurons/iaf_psc_delta0.nestml'!
[5,iaf_psc_delta0_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[6,iaf_psc_delta0_nestml, WARNING, [67:4;67:22]]: Variable 'G' has the same name as a physical unit!
[7,iaf_psc_delta0_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
[8,iaf_psc_delta0_nestml, WARNING, [69:70;69:70]]: Non-matching unit types at pA +/- pA buffer! Implicitly replaced by pA +/- 1.0 * pA buffer.
[9,iaf_psc_delta0_nestml, WARNING, [69:13;69:65]]: Non-matching unit types at mV / ms +/- pA / pF! Implicitly replaced by mV / ms +/- 1.0 * pA / pF.
[10,iaf_psc_delta0_nestml, INFO, [63:4;63:17]]: Ode of 'V_abs' updated!
[11,GLOBAL, INFO]: Start processing '/home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/synapses/stdp_triplet0.nestml'!
[12,stdp_triplet0_nestml, INFO, [2:0;67:0]]: Start building symbol table!
[13,stdp_triplet0_nestml, INFO, [64:18;64:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[14,stdp_triplet0_nestml, INFO, [46:13;46:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[15,stdp_triplet0_nestml, INFO, [47:13;47:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[16,stdp_triplet0_nestml, WARNING, [50:12;50:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[17,stdp_triplet0_nestml, INFO, [56:13;56:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[18,stdp_triplet0_nestml, INFO, [57:13;57:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[19,stdp_triplet0_nestml, WARNING, [60:12;60:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[20,GLOBAL, INFO]: All variables defined in state block: w, tr_r1, tr_r2, tr_o1, tr_o2
[21,GLOBAL, INFO]: All variables due to convolutions: []
[22,GLOBAL, INFO]: Assigned-to variables in onReceive(pre_spikes): tr_r1, tr_r2, w
[23,GLOBAL, INFO]: Variables used in convolve with other than 'spike post' port: []
[24,GLOBAL, INFO]: --> State variables that will be moved from synapse to neuron: {'tr_o2', 'tr_o1'}
[25,GLOBAL, INFO]: recursive dependent variables search yielded the following new variables: {'tr_o2', 'tr_o1', 'tau_minus', 'tau_y'}
[26,GLOBAL, INFO]: Moving state var definition of tr_o2 from synapse to neuron
[27,GLOBAL, INFO]: Moving state var definition of tr_o1 from synapse to neuron
[28,GLOBAL, INFO]: Moving state var defining equation(s) tr_o2
[29,GLOBAL, INFO]: Moving state var defining equation(s) tr_o1
[30,GLOBAL, INFO]: Moving state variables for equation(s) tr_o2
[31,GLOBAL, INFO]: Moving state variables for equation(s) tr_o1
[32,GLOBAL, INFO]: Moving onPost updates for tr_o2
[33,GLOBAL, INFO]: Moving state var updates for tr_o2 from synapse to neuron
[34,GLOBAL, INFO]: Moving onPost updates for tr_o1
[35,GLOBAL, INFO]: Moving state var updates for tr_o1 from synapse to neuron
[36,GLOBAL, INFO]: Dependent variables: tau_y, tr_o2, tau_minus, tr_o1
[37,GLOBAL, INFO]: Copying declarations from neuron equations block to synapse equations block...
[38,GLOBAL, INFO]: • Copying variable tau_y
[39,GLOBAL, INFO]: • Copying variable tr_o2
[40,GLOBAL, INFO]: • Copying variable tau_minus
[41,GLOBAL, INFO]: • Copying variable tr_o1
[42,GLOBAL, INFO]: In synapse: replacing variables with suffixed external variable references
[43,GLOBAL, INFO]: • Replacing variable tr_o2
[44,GLOBAL, INFO]: ASTSimpleExpression replacement made (var = tr_o2__for_stdp_triplet0_nestml) in expression: A3_plus * tr_o2
[45,GLOBAL, INFO]: • Replacing variable tr_o1
[46,GLOBAL, INFO]: ASTSimpleExpression replacement made (var = tr_o1__for_stdp_triplet0_nestml) in expression: tr_o1 * (A2_minus + A3_minus * tr_r2)
[47,GLOBAL, INFO]: In synapse: replacing ``continuous`` type input ports that are connected to postsynaptic neuron with suffixed external variable references
[48,GLOBAL, INFO]: Add suffix to moved variable names in neuron...
[49,GLOBAL, INFO]: • Variable tau_y
[50,GLOBAL, INFO]: • Variable tr_o2
[51,GLOBAL, INFO]: • Variable tau_minus
[52,GLOBAL, INFO]: • Variable tr_o1
[53,GLOBAL, INFO]: Moving parameters...
[54,GLOBAL, INFO]: Moving parameter with name tr_o2 from synapse to neuron
[55,GLOBAL, INFO]: Moving parameter with name tr_o1 from synapse to neuron
[56,GLOBAL, INFO]: Moving parameter with name tau_minus from synapse to neuron
[57,GLOBAL, INFO]: Moving parameter with name tau_y from synapse to neuron
[58,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[59,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, WARNING, [67:4;67:22]]: Variable 'G' has the same name as a physical unit!
[60,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
[61,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, WARNING, [69:70;69:70]]: Non-matching unit types at pA +/- pA buffer! Implicitly replaced by pA +/- 1.0 * pA buffer.
[62,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, WARNING, [69:13;69:65]]: Non-matching unit types at mV / ms +/- pA / pF! Implicitly replaced by mV / ms +/- 1.0 * pA / pF.
[63,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [63:4;63:17]]: Ode of 'V_abs' updated!
[64,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [10:4;10:17]]: Ode of 'tr_o2__for_stdp_triplet0_nestml' updated!
[65,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [9:4;9:17]]: Ode of 'tr_o1__for_stdp_triplet0_nestml' updated!
[66,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [2:0;67:0]]: Start building symbol table!
[67,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [64:18;64:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_syn": "2",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
[68,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, WARNING, [50:12;50:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[69,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [56:13;56:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[70,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [57:13;57:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[71,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, WARNING, [60:12;60:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[72,GLOBAL, INFO]: Successfully constructed neuron-synapse pair models
[73,GLOBAL, INFO]: Analysing/transforming neuron 'iaf_psc_delta0_nestml'
[74,iaf_psc_delta0_nestml, INFO, [58:0;131:0]]: Starts processing of the model 'iaf_psc_delta0_nestml'
[75,iaf_psc_delta0_nestml, INFO, [58:0;131:0]]: The neuron 'iaf_psc_delta0_nestml' will be analysed!
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Dependency analysis...
INFO:Generating numerical solver for the following symbols: V_abs
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"V_abs": "0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m"
}
}
]
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_syn": "2",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Dependency analysis...
INFO:Generating numerical solver for the following symbols: V_abs
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"V_abs": "0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m"
}
}
]
[76,iaf_psc_delta0_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[77,iaf_psc_delta0_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
},
{
"expression": "tr_o2__for_stdp_triplet0_nestml' = -tr_o2__for_stdp_triplet0_nestml / tau_y__for_stdp_triplet0_nestml",
"initial_values": {
"tr_o2__for_stdp_triplet0_nestml": "0.000000E+00"
}
},
{
"expression": "tr_o1__for_stdp_triplet0_nestml' = -tr_o1__for_stdp_triplet0_nestml / tau_minus__for_stdp_triplet0_nestml",
"initial_values": {
"tr_o1__for_stdp_triplet0_nestml": "0.000000E+00"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_minus__for_stdp_triplet0_nestml": "3.370000E+01",
"tau_syn": "2",
"tau_y__for_stdp_triplet0_nestml": "125",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet0_nestml with defining expression = "-tr_o2__for_stdp_triplet0_nestml / tau_y__for_stdp_triplet0_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet0_nestml with defining expression = "-tr_o1__for_stdp_triplet0_nestml / tau_minus__for_stdp_triplet0_nestml"
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet0_nestml with defining expression = "-tr_o2__for_stdp_triplet0_nestml / tau_y__for_stdp_triplet0_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet0_nestml with defining expression = "-tr_o1__for_stdp_triplet0_nestml / tau_minus__for_stdp_triplet0_nestml"
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Shape tr_o2__for_stdp_triplet0_nestml: reconstituting expression -tr_o2__for_stdp_triplet0_nestml/tau_y__for_stdp_triplet0_nestml
INFO:Shape tr_o1__for_stdp_triplet0_nestml: reconstituting expression -tr_o1__for_stdp_triplet0_nestml/tau_minus__for_stdp_triplet0_nestml
INFO:Dependency analysis...
INFO:Generating propagators for the following symbols: tr_o2__for_stdp_triplet0_nestml, tr_o1__for_stdp_triplet0_nestml
[78,GLOBAL, INFO]: Analysing/transforming neuron 'iaf_psc_delta0_nestml__with_stdp_triplet0_nestml'
[79,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [58:0;131:0]]: Starts processing of the model 'iaf_psc_delta0_nestml__with_stdp_triplet0_nestml'
[80,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [58:0;131:0]]: The neuron 'iaf_psc_delta0_nestml__with_stdp_triplet0_nestml' will be analysed!
INFO:Generating numerical solver for the following symbols: V_abs
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"tr_o1__for_stdp_triplet0_nestml": "0.0",
"tr_o2__for_stdp_triplet0_nestml": "0.0"
},
"parameters": {
"tau_minus__for_stdp_triplet0_nestml": "33.7000000000000",
"tau_y__for_stdp_triplet0_nestml": "125.000000000000"
},
"propagators": {
"__P__tr_o1__for_stdp_triplet0_nestml__tr_o1__for_stdp_triplet0_nestml": "exp(-__h/tau_minus__for_stdp_triplet0_nestml)",
"__P__tr_o2__for_stdp_triplet0_nestml__tr_o2__for_stdp_triplet0_nestml": "exp(-__h/tau_y__for_stdp_triplet0_nestml)"
},
"solver": "analytical",
"state_variables": [
"tr_o2__for_stdp_triplet0_nestml",
"tr_o1__for_stdp_triplet0_nestml"
],
"update_expressions": {
"tr_o1__for_stdp_triplet0_nestml": "__P__tr_o1__for_stdp_triplet0_nestml__tr_o1__for_stdp_triplet0_nestml*tr_o1__for_stdp_triplet0_nestml",
"tr_o2__for_stdp_triplet0_nestml": "__P__tr_o2__for_stdp_triplet0_nestml__tr_o2__for_stdp_triplet0_nestml*tr_o2__for_stdp_triplet0_nestml"
}
},
{
"initial_values": {
"V_abs": "0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m"
}
}
]
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
},
{
"expression": "tr_o2__for_stdp_triplet0_nestml' = -tr_o2__for_stdp_triplet0_nestml / tau_y__for_stdp_triplet0_nestml",
"initial_values": {
"tr_o2__for_stdp_triplet0_nestml": "0.000000E+00"
}
},
{
"expression": "tr_o1__for_stdp_triplet0_nestml' = -tr_o1__for_stdp_triplet0_nestml / tau_minus__for_stdp_triplet0_nestml",
"initial_values": {
"tr_o1__for_stdp_triplet0_nestml": "0.000000E+00"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_minus__for_stdp_triplet0_nestml": "3.370000E+01",
"tau_syn": "2",
"tau_y__for_stdp_triplet0_nestml": "125",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet0_nestml with defining expression = "-tr_o2__for_stdp_triplet0_nestml / tau_y__for_stdp_triplet0_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet0_nestml with defining expression = "-tr_o1__for_stdp_triplet0_nestml / tau_minus__for_stdp_triplet0_nestml"
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet0_nestml with defining expression = "-tr_o2__for_stdp_triplet0_nestml / tau_y__for_stdp_triplet0_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet0_nestml with defining expression = "-tr_o1__for_stdp_triplet0_nestml / tau_minus__for_stdp_triplet0_nestml"
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Shape tr_o2__for_stdp_triplet0_nestml: reconstituting expression -tr_o2__for_stdp_triplet0_nestml/tau_y__for_stdp_triplet0_nestml
INFO:Shape tr_o1__for_stdp_triplet0_nestml: reconstituting expression -tr_o1__for_stdp_triplet0_nestml/tau_minus__for_stdp_triplet0_nestml
INFO:Dependency analysis...
INFO:Generating numerical solver for the following symbols: V_abs, tr_o2__for_stdp_triplet0_nestml, tr_o1__for_stdp_triplet0_nestml
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"V_abs": "0",
"tr_o1__for_stdp_triplet0_nestml": "0.0",
"tr_o2__for_stdp_triplet0_nestml": "0.0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000",
"tau_minus__for_stdp_triplet0_nestml": "33.7000000000000",
"tau_y__for_stdp_triplet0_nestml": "125.000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs",
"tr_o2__for_stdp_triplet0_nestml",
"tr_o1__for_stdp_triplet0_nestml"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m",
"tr_o1__for_stdp_triplet0_nestml": "-tr_o1__for_stdp_triplet0_nestml/tau_minus__for_stdp_triplet0_nestml",
"tr_o2__for_stdp_triplet0_nestml": "-tr_o2__for_stdp_triplet0_nestml/tau_y__for_stdp_triplet0_nestml"
}
}
]
[81,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[82,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "tr_r1' = -tr_r1 / tau_plus",
"initial_values": {
"tr_r1": "0.000000E+00"
}
},
{
"expression": "tr_r2' = -tr_r2 / tau_x",
"initial_values": {
"tr_r2": "0.000000E+00"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"A2_minus": "7.000000E-03",
"A2_plus": "7.500000E-10",
"A3_minus": "2.300000E-04",
"A3_plus": "9.300000E-03",
"Wmax": "100",
"Wmin": "0",
"tau_plus": "1.680000E+01",
"tau_x": "101",
"the_delay": "1"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape tr_r1 with defining expression = "-tr_r1 / tau_plus"
INFO:
Processing shape tr_r2 with defining expression = "-tr_r2 / tau_x"
INFO:
Processing shape tr_r1 with defining expression = "-tr_r1 / tau_plus"
INFO:
Processing shape tr_r2 with defining expression = "-tr_r2 / tau_x"
INFO:Shape tr_r1: reconstituting expression -tr_r1/tau_plus
INFO:Shape tr_r2: reconstituting expression -tr_r2/tau_x
INFO:Dependency analysis...
INFO:Generating propagators for the following symbols: tr_r1, tr_r2
Analysing/transforming synapse stdp_triplet0_nestml__with_iaf_psc_delta0_nestml.
[83,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [2:0;67:0]]: Starts processing of the model 'stdp_triplet0_nestml__with_iaf_psc_delta0_nestml'
[84,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [2:0;67:0]]: The neuron 'stdp_triplet0_nestml__with_iaf_psc_delta0_nestml' will be analysed!
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"tr_r1": "0.0",
"tr_r2": "0.0"
},
"parameters": {
"tau_plus": "16.8000000000000",
"tau_x": "101.000000000000"
},
"propagators": {
"__P__tr_r1__tr_r1": "exp(-__h/tau_plus)",
"__P__tr_r2__tr_r2": "exp(-__h/tau_x)"
},
"solver": "analytical",
"state_variables": [
"tr_r1",
"tr_r2"
],
"update_expressions": {
"tr_r1": "__P__tr_r1__tr_r1*tr_r1",
"tr_r2": "__P__tr_r2__tr_r2*tr_r2"
}
}
]
[85,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [2:0;67:0]]: Start building symbol table!
[86,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [64:18;64:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[87,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [56:13;56:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[88,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [57:13;57:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[89,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [2:0;67:0]]: Start building symbol table!
[90,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [64:18;64:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[91,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [56:13;56:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[92,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [57:13;57:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[93,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [2:0;67:0]]: Start building symbol table!
[94,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [64:18;64:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[95,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [56:13;56:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[96,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [57:13;57:13]]: Implicit casting from (compatible) type 'integer' to 'real'.
[97,iaf_psc_delta0_nestml, INFO, [58:0;131:0]]: Successfully generated code for the model: 'iaf_psc_delta0_nestml' in: '/tmp/nestml_module' !
[98,iaf_psc_delta0_nestml__with_stdp_triplet0_nestml, INFO, [58:0;131:0]]: Successfully generated code for the model: 'iaf_psc_delta0_nestml__with_stdp_triplet0_nestml' in: '/tmp/nestml_module' !
Generating code for the synapse stdp_triplet0_nestml__with_iaf_psc_delta0_nestml.
[99,stdp_triplet0_nestml__with_iaf_psc_delta0_nestml, INFO, [2:0;67:0]]: Successfully generated code for the model: 'stdp_triplet0_nestml__with_iaf_psc_delta0_nestml' in: '/tmp/nestml_module' !
[100,GLOBAL, INFO]: Successfully generated NEST module code in '/tmp/nestml_module' !
\(\textit{Nearest spike Interation}\) - Each postsynaptic spike interacts with only the last spike. Hence no accumulation of the trace variables occurs. The variables saturate at 1. This is achieved by updating the variables to the value of 1 instead of updating by at step of 1. In this way, the synapse forgets all other previous spikes and keeps only the memory of the last one.
[6]:
nestml_triplet_stdp_nn_model = '''
synapse stdp_triplet_nn:
state:
w nS = 1 nS
tr_r1 real = 0.
tr_r2 real = 0.
tr_o1 real = 0.
tr_o2 real = 0.
end
parameters:
the_delay ms = 1 ms @nest::delay # !!! cannot have a variable called "delay"
tau_plus ms = 16.8 ms # time constant for tr_r1
tau_x ms = 101 ms # time constant for tr_r2
tau_minus ms = 33.7 ms # time constant for tr_o1
tau_y ms = 125 ms # time constant for tr_o2
A2_plus real = 7.5e-10
A3_plus real = 9.3e-3
A2_minus real = 7e-3
A3_minus real = 2.3e-4
Wmax nS = 100 nS
Wmin nS = 0 nS
end
equations:
tr_r1' = -tr_r1 / tau_plus
tr_r2' = -tr_r2 / tau_x
tr_o1' = -tr_o1 / tau_minus
tr_o2' = -tr_o2 / tau_y
end
input:
pre_spikes nS <- spike
post_spikes nS <- spike
end
output: spike
onReceive(post_spikes):
# increment post trace values
tr_o1 = 1
tr_o2 = 1
# potentiate synapse
#w_ nS = Wmax * ( w / Wmax + tr_r1 * ( A2_plus + A3_plus * tr_o2 ) )
w_ nS = w + tr_r1 * ( A2_plus + A3_plus * tr_o2 )
w = min(Wmax, w_)
end
onReceive(pre_spikes):
# increment pre trace values
tr_r1 = 1
tr_r2 = 1
# depress synapse
#w_ nS = Wmax * ( w / Wmax - tr_o1 * ( A2_minus + A3_minus * tr_r2 ) )
w_ nS = w - tr_o1 * ( A2_minus + A3_minus * tr_r2 )
w = max(Wmin, w_)
# deliver spike to postsynaptic partner
deliver_spike(w, the_delay)
end
end
'''
[7]:
# Generate code for nearest spike interaction model
neuron_model_name_nn, synapse_model_name_nn, nestml_synapse_model_name_nn = \
generate_code_for(nestml_triplet_stdp_nn_model)
[1,GLOBAL, INFO]: List of files that will be processed:
[2,GLOBAL, INFO]: /home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/neurons/iaf_psc_delta1.nestml
[3,GLOBAL, INFO]: /home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/synapses/stdp_triplet_nn1.nestml
[4,GLOBAL, INFO]: Start processing '/home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/neurons/iaf_psc_delta1.nestml'!
[5,iaf_psc_delta1_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[6,iaf_psc_delta1_nestml, WARNING, [67:4;67:22]]: Variable 'G' has the same name as a physical unit!
[7,iaf_psc_delta1_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
[8,iaf_psc_delta1_nestml, WARNING, [69:70;69:70]]: Non-matching unit types at pA +/- pA buffer! Implicitly replaced by pA +/- 1.0 * pA buffer.
[9,iaf_psc_delta1_nestml, WARNING, [69:13;69:65]]: Non-matching unit types at mV / ms +/- pA / pF! Implicitly replaced by mV / ms +/- 1.0 * pA / pF.
[10,iaf_psc_delta1_nestml, INFO, [63:4;63:17]]: Ode of 'V_abs' updated!
[11,GLOBAL, INFO]: Start processing '/home/charl/julich/nestml-fork-jit-third-factor/nestml/doc/tutorials/stdp_windows/models/synapses/stdp_triplet_nn1.nestml'!
[12,stdp_triplet_nn1_nestml, INFO, [2:0;69:0]]: Start building symbol table!
[13,stdp_triplet_nn1_nestml, INFO, [66:18;66:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[14,stdp_triplet_nn1_nestml, INFO, [46:12;46:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[15,stdp_triplet_nn1_nestml, INFO, [47:12;47:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[16,stdp_triplet_nn1_nestml, WARNING, [51:12;51:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[17,stdp_triplet_nn1_nestml, INFO, [57:12;57:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[18,stdp_triplet_nn1_nestml, INFO, [58:12;58:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[19,stdp_triplet_nn1_nestml, WARNING, [62:12;62:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[20,GLOBAL, INFO]: All variables defined in state block: w, tr_r1, tr_r2, tr_o1, tr_o2
[21,GLOBAL, INFO]: All variables due to convolutions: []
[22,GLOBAL, INFO]: Assigned-to variables in onReceive(pre_spikes): tr_r1, tr_r2, w
[23,GLOBAL, INFO]: Variables used in convolve with other than 'spike post' port: []
[24,GLOBAL, INFO]: --> State variables that will be moved from synapse to neuron: {'tr_o2', 'tr_o1'}
[25,GLOBAL, INFO]: recursive dependent variables search yielded the following new variables: {'tr_o2', 'tr_o1', 'tau_minus', 'tau_y'}
[26,GLOBAL, INFO]: Moving state var definition of tr_o2 from synapse to neuron
[27,GLOBAL, INFO]: Moving state var definition of tr_o1 from synapse to neuron
[28,GLOBAL, INFO]: Moving state var defining equation(s) tr_o2
[29,GLOBAL, INFO]: Moving state var defining equation(s) tr_o1
[30,GLOBAL, INFO]: Moving state variables for equation(s) tr_o2
[31,GLOBAL, INFO]: Moving state variables for equation(s) tr_o1
[32,GLOBAL, INFO]: Moving onPost updates for tr_o2
[33,GLOBAL, INFO]: Moving state var updates for tr_o2 from synapse to neuron
[34,GLOBAL, INFO]: Moving onPost updates for tr_o1
[35,GLOBAL, INFO]: Moving state var updates for tr_o1 from synapse to neuron
[36,GLOBAL, INFO]: Dependent variables: tau_y, tr_o2, tr_o1, tau_minus
[37,GLOBAL, INFO]: Copying declarations from neuron equations block to synapse equations block...
[38,GLOBAL, INFO]: • Copying variable tau_y
[39,GLOBAL, INFO]: • Copying variable tr_o2
[40,GLOBAL, INFO]: • Copying variable tr_o1
[41,GLOBAL, INFO]: • Copying variable tau_minus
[42,GLOBAL, INFO]: In synapse: replacing variables with suffixed external variable references
[43,GLOBAL, INFO]: • Replacing variable tr_o2
[44,GLOBAL, INFO]: ASTSimpleExpression replacement made (var = tr_o2__for_stdp_triplet_nn1_nestml) in expression: A3_plus * tr_o2
[45,GLOBAL, INFO]: • Replacing variable tr_o1
[46,GLOBAL, INFO]: ASTSimpleExpression replacement made (var = tr_o1__for_stdp_triplet_nn1_nestml) in expression: tr_o1 * (A2_minus + A3_minus * tr_r2)
[47,GLOBAL, INFO]: In synapse: replacing ``continuous`` type input ports that are connected to postsynaptic neuron with suffixed external variable references
[48,GLOBAL, INFO]: Add suffix to moved variable names in neuron...
[49,GLOBAL, INFO]: • Variable tau_y
[50,GLOBAL, INFO]: • Variable tr_o2
[51,GLOBAL, INFO]: • Variable tr_o1
[52,GLOBAL, INFO]: • Variable tau_minus
[53,GLOBAL, INFO]: Moving parameters...
[54,GLOBAL, INFO]: Moving parameter with name tr_o2 from synapse to neuron
[55,GLOBAL, INFO]: Moving parameter with name tr_o1 from synapse to neuron
[56,GLOBAL, INFO]: Moving parameter with name tau_minus from synapse to neuron
[57,GLOBAL, INFO]: Moving parameter with name tau_y from synapse to neuron
[58,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[59,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, WARNING, [67:4;67:22]]: Variable 'G' has the same name as a physical unit!
[60,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
[61,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, WARNING, [69:70;69:70]]: Non-matching unit types at pA +/- pA buffer! Implicitly replaced by pA +/- 1.0 * pA buffer.
[62,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, WARNING, [69:13;69:65]]: Non-matching unit types at mV / ms +/- pA / pF! Implicitly replaced by mV / ms +/- 1.0 * pA / pF.
[63,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [63:4;63:17]]: Ode of 'V_abs' updated!
[64,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [10:4;10:17]]: Ode of 'tr_o2__for_stdp_triplet_nn1_nestml' updated!
[65,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [9:4;9:17]]: Ode of 'tr_o1__for_stdp_triplet_nn1_nestml' updated!
[66,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [2:0;69:0]]: Start building symbol table!
[67,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [66:18;66:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_syn": "2",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
[68,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, WARNING, [51:12;51:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[69,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [57:12;57:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[70,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [58:12;58:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[71,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, WARNING, [62:12;62:12]]: Implicit casting from (compatible) type 'nS' to 'real'.
[72,GLOBAL, INFO]: Successfully constructed neuron-synapse pair models
[73,GLOBAL, INFO]: Analysing/transforming neuron 'iaf_psc_delta1_nestml'
[74,iaf_psc_delta1_nestml, INFO, [58:0;131:0]]: Starts processing of the model 'iaf_psc_delta1_nestml'
[75,iaf_psc_delta1_nestml, INFO, [58:0;131:0]]: The neuron 'iaf_psc_delta1_nestml' will be analysed!
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Dependency analysis...
INFO:Generating numerical solver for the following symbols: V_abs
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"V_abs": "0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m"
}
}
]
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_syn": "2",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Dependency analysis...
INFO:Generating numerical solver for the following symbols: V_abs
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"V_abs": "0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m"
}
}
]
[76,iaf_psc_delta1_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[77,iaf_psc_delta1_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
},
{
"expression": "tr_o2__for_stdp_triplet_nn1_nestml' = -tr_o2__for_stdp_triplet_nn1_nestml / tau_y__for_stdp_triplet_nn1_nestml",
"initial_values": {
"tr_o2__for_stdp_triplet_nn1_nestml": "0.000000E+00"
}
},
{
"expression": "tr_o1__for_stdp_triplet_nn1_nestml' = -tr_o1__for_stdp_triplet_nn1_nestml / tau_minus__for_stdp_triplet_nn1_nestml",
"initial_values": {
"tr_o1__for_stdp_triplet_nn1_nestml": "0.000000E+00"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_minus__for_stdp_triplet_nn1_nestml": "3.370000E+01",
"tau_syn": "2",
"tau_y__for_stdp_triplet_nn1_nestml": "125",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o2__for_stdp_triplet_nn1_nestml / tau_y__for_stdp_triplet_nn1_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o1__for_stdp_triplet_nn1_nestml / tau_minus__for_stdp_triplet_nn1_nestml"
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o2__for_stdp_triplet_nn1_nestml / tau_y__for_stdp_triplet_nn1_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o1__for_stdp_triplet_nn1_nestml / tau_minus__for_stdp_triplet_nn1_nestml"
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Shape tr_o2__for_stdp_triplet_nn1_nestml: reconstituting expression -tr_o2__for_stdp_triplet_nn1_nestml/tau_y__for_stdp_triplet_nn1_nestml
INFO:Shape tr_o1__for_stdp_triplet_nn1_nestml: reconstituting expression -tr_o1__for_stdp_triplet_nn1_nestml/tau_minus__for_stdp_triplet_nn1_nestml
INFO:Dependency analysis...
INFO:Generating propagators for the following symbols: tr_o2__for_stdp_triplet_nn1_nestml, tr_o1__for_stdp_triplet_nn1_nestml
[78,GLOBAL, INFO]: Analysing/transforming neuron 'iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml'
[79,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [58:0;131:0]]: Starts processing of the model 'iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml'
[80,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [58:0;131:0]]: The neuron 'iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml' will be analysed!
INFO:Generating numerical solver for the following symbols: V_abs
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"tr_o1__for_stdp_triplet_nn1_nestml": "0.0",
"tr_o2__for_stdp_triplet_nn1_nestml": "0.0"
},
"parameters": {
"tau_minus__for_stdp_triplet_nn1_nestml": "33.7000000000000",
"tau_y__for_stdp_triplet_nn1_nestml": "125.000000000000"
},
"propagators": {
"__P__tr_o1__for_stdp_triplet_nn1_nestml__tr_o1__for_stdp_triplet_nn1_nestml": "exp(-__h/tau_minus__for_stdp_triplet_nn1_nestml)",
"__P__tr_o2__for_stdp_triplet_nn1_nestml__tr_o2__for_stdp_triplet_nn1_nestml": "exp(-__h/tau_y__for_stdp_triplet_nn1_nestml)"
},
"solver": "analytical",
"state_variables": [
"tr_o2__for_stdp_triplet_nn1_nestml",
"tr_o1__for_stdp_triplet_nn1_nestml"
],
"update_expressions": {
"tr_o1__for_stdp_triplet_nn1_nestml": "__P__tr_o1__for_stdp_triplet_nn1_nestml__tr_o1__for_stdp_triplet_nn1_nestml*tr_o1__for_stdp_triplet_nn1_nestml",
"tr_o2__for_stdp_triplet_nn1_nestml": "__P__tr_o2__for_stdp_triplet_nn1_nestml__tr_o2__for_stdp_triplet_nn1_nestml*tr_o2__for_stdp_triplet_nn1_nestml"
}
},
{
"initial_values": {
"V_abs": "0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m"
}
}
]
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "V_abs' = -V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m",
"initial_values": {
"V_abs": "0"
}
},
{
"expression": "tr_o2__for_stdp_triplet_nn1_nestml' = -tr_o2__for_stdp_triplet_nn1_nestml / tau_y__for_stdp_triplet_nn1_nestml",
"initial_values": {
"tr_o2__for_stdp_triplet_nn1_nestml": "0.000000E+00"
}
},
{
"expression": "tr_o1__for_stdp_triplet_nn1_nestml' = -tr_o1__for_stdp_triplet_nn1_nestml / tau_minus__for_stdp_triplet_nn1_nestml",
"initial_values": {
"tr_o1__for_stdp_triplet_nn1_nestml": "0.000000E+00"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"C_m": "250",
"E_L": "-70",
"I_e": "0",
"Theta": "-55 - E_L",
"V_min": "-inf * 1",
"V_reset": "-70 - E_L",
"t_ref": "2",
"tau_m": "10",
"tau_minus__for_stdp_triplet_nn1_nestml": "3.370000E+01",
"tau_syn": "2",
"tau_y__for_stdp_triplet_nn1_nestml": "125",
"with_refr_input": "false"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o2__for_stdp_triplet_nn1_nestml / tau_y__for_stdp_triplet_nn1_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o1__for_stdp_triplet_nn1_nestml / tau_minus__for_stdp_triplet_nn1_nestml"
INFO:
Processing shape V_abs with defining expression = "-V_abs / tau_m + (1.0 / 1.0 / 1.0) * 0 + (I_e + I_stim) / C_m"
INFO:
Processing shape tr_o2__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o2__for_stdp_triplet_nn1_nestml / tau_y__for_stdp_triplet_nn1_nestml"
INFO:
Processing shape tr_o1__for_stdp_triplet_nn1_nestml with defining expression = "-tr_o1__for_stdp_triplet_nn1_nestml / tau_minus__for_stdp_triplet_nn1_nestml"
INFO:Shape V_abs: reconstituting expression -V_abs/tau_m + (I_e + I_stim)/C_m
INFO:Shape tr_o2__for_stdp_triplet_nn1_nestml: reconstituting expression -tr_o2__for_stdp_triplet_nn1_nestml/tau_y__for_stdp_triplet_nn1_nestml
INFO:Shape tr_o1__for_stdp_triplet_nn1_nestml: reconstituting expression -tr_o1__for_stdp_triplet_nn1_nestml/tau_minus__for_stdp_triplet_nn1_nestml
INFO:Dependency analysis...
INFO:Generating numerical solver for the following symbols: V_abs, tr_o2__for_stdp_triplet_nn1_nestml, tr_o1__for_stdp_triplet_nn1_nestml
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"V_abs": "0",
"tr_o1__for_stdp_triplet_nn1_nestml": "0.0",
"tr_o2__for_stdp_triplet_nn1_nestml": "0.0"
},
"parameters": {
"C_m": "250.000000000000",
"I_e": "0",
"tau_m": "10.0000000000000",
"tau_minus__for_stdp_triplet_nn1_nestml": "33.7000000000000",
"tau_y__for_stdp_triplet_nn1_nestml": "125.000000000000"
},
"solver": "numeric",
"state_variables": [
"V_abs",
"tr_o2__for_stdp_triplet_nn1_nestml",
"tr_o1__for_stdp_triplet_nn1_nestml"
],
"update_expressions": {
"V_abs": "-V_abs/tau_m + (I_e + I_stim)/C_m",
"tr_o1__for_stdp_triplet_nn1_nestml": "-tr_o1__for_stdp_triplet_nn1_nestml/tau_minus__for_stdp_triplet_nn1_nestml",
"tr_o2__for_stdp_triplet_nn1_nestml": "-tr_o2__for_stdp_triplet_nn1_nestml/tau_y__for_stdp_triplet_nn1_nestml"
}
}
]
[81,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [58:0;131:0]]: Start building symbol table!
[82,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, WARNING, [88:4;88:22]]: Variable 'h' has the same name as a physical unit!
INFO:ode-toolbox: analysing input:
INFO:{
"dynamics": [
{
"expression": "tr_r1' = -tr_r1 / tau_plus",
"initial_values": {
"tr_r1": "0.000000E+00"
}
},
{
"expression": "tr_r2' = -tr_r2 / tau_x",
"initial_values": {
"tr_r2": "0.000000E+00"
}
}
],
"options": {
"output_timestep_symbol": "__h"
},
"parameters": {
"A2_minus": "7.000000E-03",
"A2_plus": "7.500000E-10",
"A3_minus": "2.300000E-04",
"A3_plus": "9.300000E-03",
"Wmax": "100",
"Wmin": "0",
"tau_plus": "1.680000E+01",
"tau_x": "101",
"the_delay": "1"
}
}
INFO:Processing global options...
INFO:Processing input shapes...
INFO:
Processing shape tr_r1 with defining expression = "-tr_r1 / tau_plus"
INFO:
Processing shape tr_r2 with defining expression = "-tr_r2 / tau_x"
INFO:
Processing shape tr_r1 with defining expression = "-tr_r1 / tau_plus"
INFO:
Processing shape tr_r2 with defining expression = "-tr_r2 / tau_x"
INFO:Shape tr_r1: reconstituting expression -tr_r1/tau_plus
INFO:Shape tr_r2: reconstituting expression -tr_r2/tau_x
INFO:Dependency analysis...
INFO:Generating propagators for the following symbols: tr_r1, tr_r2
Analysing/transforming synapse stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml.
[83,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [2:0;69:0]]: Starts processing of the model 'stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml'
[84,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [2:0;69:0]]: The neuron 'stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml' will be analysed!
INFO:In ode-toolbox: returning outdict =
INFO:[
{
"initial_values": {
"tr_r1": "0.0",
"tr_r2": "0.0"
},
"parameters": {
"tau_plus": "16.8000000000000",
"tau_x": "101.000000000000"
},
"propagators": {
"__P__tr_r1__tr_r1": "exp(-__h/tau_plus)",
"__P__tr_r2__tr_r2": "exp(-__h/tau_x)"
},
"solver": "analytical",
"state_variables": [
"tr_r1",
"tr_r2"
],
"update_expressions": {
"tr_r1": "__P__tr_r1__tr_r1*tr_r1",
"tr_r2": "__P__tr_r2__tr_r2*tr_r2"
}
}
]
[85,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [2:0;69:0]]: Start building symbol table!
[86,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [66:18;66:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[87,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [57:12;57:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[88,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [58:12;58:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[89,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [2:0;69:0]]: Start building symbol table!
[90,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [66:18;66:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[91,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [57:12;57:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[92,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [58:12;58:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[93,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [2:0;69:0]]: Start building symbol table!
[94,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [66:18;66:18]]: Implicit casting from (compatible) type 'nS' to 'real'.
[95,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [57:12;57:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[96,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [58:12;58:12]]: Implicit casting from (compatible) type 'integer' to 'real'.
[97,iaf_psc_delta1_nestml, INFO, [58:0;131:0]]: Successfully generated code for the model: 'iaf_psc_delta1_nestml' in: '/tmp/nestml_module' !
[98,iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml, INFO, [58:0;131:0]]: Successfully generated code for the model: 'iaf_psc_delta1_nestml__with_stdp_triplet_nn1_nestml' in: '/tmp/nestml_module' !
Generating code for the synapse stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml.
[99,stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml, INFO, [2:0;69:0]]: Successfully generated code for the model: 'stdp_triplet_nn1_nestml__with_iaf_psc_delta1_nestml' in: '/tmp/nestml_module' !
[100,GLOBAL, INFO]: Successfully generated NEST module code in '/tmp/nestml_module' !
Results
Spike pairing protocol
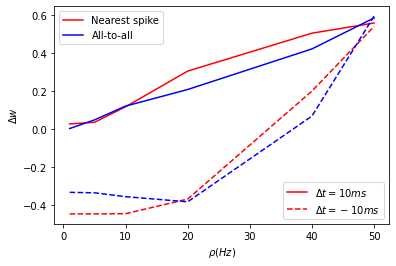
If we set the values of \(A_3^+\) and \(A_3^-\) to 0 in the above equations, the model becomes a classical STDP model. The authors in [4] tested this model to check if they could reproduce the experimental results of [2] and [3]. They tested this with a pairing protocol which is a classic STDP protocol where the pairs of presynaptic and postsynaptic spikes shifted by \(\Delta{t}\) are repeated at regular intervals of \((1/\rho)\). They showed that spike pairs repeated at high frequencies did not have considerable effect on the synaptic weights as shown by the experimental data.
Triplet STDP model with spike pairs
Let us simulate the pairing protocol in the above formulated model to see if we can reproduce the frequency dependence of spikes on the synaptic weights.
[8]:
# Create pre and post spike arrays
def create_spike_pairs(freq=1., delta_t=0., size=10):
"""
Creates the spike pairs given the frequency and the time difference between the pre and post spikes.
:param: freq: frequency of the spike pairs
:param: delta_t: time difference or delay between the spikes
:param: size: number of spike pairs to be generated.
:return: pre and post spike arrays
"""
pre_spike_times = 1 + abs(delta_t) + freq * np.arange(size).astype(float)
post_spike_times = 1 + abs(delta_t) + delta_t + freq * np.arange(size).astype(float)
return pre_spike_times, post_spike_times
[9]:
def run_triplet_stdp_network(neuron_model_name, synapse_model_name, neuron_opts,
nest_syn_opts, pre_spike_times, post_spike_times,
resolution=1., delay=1., sim_time=None):
"""
Runs the triplet stdp synapse model
"""
nest.set_verbosity("M_ALL")
nest.ResetKernel()
nest.SetKernelStatus({'resolution': resolution})
# Set defaults for neuron
nest.SetDefaults(neuron_model_name, neuron_opts)
# Create neurons
neurons = nest.Create(neuron_model_name, 2)
pre_sg = nest.Create('spike_generator', params={'spike_times': pre_spike_times})
post_sg = nest.Create('spike_generator', params={'spike_times': post_spike_times})
spikes = nest.Create('spike_recorder')
weight_recorder = nest.Create('weight_recorder')
# Set defaults for synapse
nest.CopyModel('static_synapse',
'excitatory_noise',
{'weight': 9999.,
'delay' : syn_opts["delay"]})
_syn_opts = nest_syn_opts.copy()
_syn_opts.pop('delay')
nest.CopyModel(synapse_model_name,
synapse_model_name + "_rec",
{'weight_recorder' : weight_recorder[0]})
nest.SetDefaults(synapse_model_name + "_rec", _syn_opts)
# Connect nodes
nest.Connect(neurons[0], neurons[1], syn_spec={'synapse_model': synapse_model_name + "_rec"})
nest.Connect(pre_sg, neurons[0], syn_spec='excitatory_noise')
nest.Connect(post_sg, neurons[1], syn_spec='excitatory_noise')
nest.Connect(neurons, spikes)
# Run simulation
syn = nest.GetConnections(source=neurons[0], synapse_model=synapse_model_name + "_rec")
initial_weight = nest.GetStatus(syn)[0]['w']
nest.Simulate(sim_time)
updated_weight = nest.GetStatus(syn)[0]['w']
dw = updated_weight - initial_weight
print('Initial weight: {}, Updated weight: {}'.format(initial_weight, updated_weight))
connections = nest.GetConnections(neurons, neurons)
gid_pre = nest.GetStatus(connections,'source')[0]
gid_post = nest.GetStatus(connections,'target')[0]
# From the spike recorder
events = nest.GetStatus(spikes, 'events')[0]
times_spikes = np.array(events['times'])
senders_spikes = events['senders']
# print('times_spikes: ', times_spikes)
# print('senders_spikes: ', senders_spikes)
# From the weight recorder
events = nest.GetStatus(weight_recorder, 'events')[0]
times_weights = events['times']
weights = events['weights']
# print('times_weights: ', times_weights)
# print('weights: ', weights)
return dw
Simulation
[10]:
# Simulate the network
def run_frequency_simulation(neuron_model_name, synapse_model_name,
neuron_opts, nest_syn_opts,
freqs, delta_t, n_spikes):
"""
Runs the spike pair simulation for given frequencies and given time difference between spikes.
"""
dw_dict = dict.fromkeys(delta_t)
for _t in delta_t:
dw_vec = []
for _freq in freqs:
spike_interval = (1/_freq)*1000 # in ms
pre_spike_times, post_spike_times = create_spike_pairs(freq=spike_interval,
delta_t=_t,
size=n_spikes)
sim_time = max(np.amax(pre_spike_times), np.amax(post_spike_times)) + 10. + 3 * syn_opts["delay"]
dw = run_triplet_stdp_network(neuron_model_name, synapse_model_name,
neuron_opts, nest_syn_opts,
pre_spike_times=pre_spike_times,
post_spike_times=post_spike_times,
sim_time=sim_time)
dw_vec.append(dw)
dw_dict[_t] = dw_vec
return dw_dict
All-to-all spike interaction
[35]:
freqs = [1., 5., 10., 20., 40., 50.] # frequency of spikes in Hz
delta_t = [10, -10] # delay between t_post and t_pre in ms
n_spikes = 60
[36]:
syn_opts = {
'delay': 1.,
'tau_minus': 33.7,
'tau_plus': 16.8,
'tau_x': 101.,
'tau_y': 125.,
'A2_plus': 5e-10,
'A3_plus': 6.2e-3,
'A2_minus': 7e-3,
'A3_minus': 2.3e-4,
'Wmax': 50.,
'Wmin' : 0.,
'w': 1.
}
synapse_suffix = neuron_model_name[neuron_model_name.find(nestml_synapse_model_name):]
neuron_opts = {'tau_minus__for_' + synapse_suffix: syn_opts['tau_minus'],
'tau_y__for_' + synapse_suffix: syn_opts['tau_y']}
nest_syn_opts = syn_opts.copy()
nest_syn_opts.pop('tau_minus')
nest_syn_opts.pop('tau_y')
dw_dict = run_frequency_simulation(neuron_model_name, synapse_model_name,
neuron_opts, nest_syn_opts,
freqs, delta_t, n_spikes)
Initial weight: 1.0, Updated weight: 1.0032840201153659
Initial weight: 1.0, Updated weight: 1.0487030313494627
Initial weight: 1.0, Updated weight: 1.1212921010110624
Initial weight: 1.0, Updated weight: 1.208550316936044
Initial weight: 1.0, Updated weight: 1.4218868273243082
Initial weight: 1.0, Updated weight: 1.5846034621613552
Initial weight: 1.0, Updated weight: 0.6678711978627694
Initial weight: 1.0, Updated weight: 0.6653426131462727
Initial weight: 1.0, Updated weight: 0.6450780469148971
Initial weight: 1.0, Updated weight: 0.6180411107607721
Initial weight: 1.0, Updated weight: 1.068737821702289
Initial weight: 1.0, Updated weight: 1.5937453662768748
Nearest spike interaction
[37]:
syn_opts_nn = {
'delay': 1.,
'tau_minus': 33.7,
'tau_plus': 16.8,
'tau_x': 714.,
'tau_y': 40.,
'A2_plus': 8.8e-11,
'A3_plus': 5.3e-2,
'A2_minus': 6.6e-3,
'A3_minus': 3.1e-3,
'Wmax': 50.,
'Wmin' : 0.,
'w': 1.
}
synapse_suffix_nn = neuron_model_name_nn[neuron_model_name_nn.find(nestml_synapse_model_name_nn):]
neuron_opts_nn = {'tau_minus__for_' + synapse_suffix_nn: syn_opts_nn['tau_minus'],
'tau_y__for_' + synapse_suffix_nn: syn_opts_nn['tau_y']}
nest_syn_opts_nn = syn_opts_nn.copy()
nest_syn_opts_nn.pop('tau_minus')
nest_syn_opts_nn.pop('tau_y')
dw_dict_nn = run_frequency_simulation(neuron_model_name_nn, synapse_model_name_nn,
neuron_opts_nn, nest_syn_opts_nn,
freqs, delta_t, n_spikes)
Initial weight: 1.0, Updated weight: 1.027536987681302
Initial weight: 1.0, Updated weight: 1.0361996900270407
Initial weight: 1.0, Updated weight: 1.1178373501754864
Initial weight: 1.0, Updated weight: 1.3052281386777107
Initial weight: 1.0, Updated weight: 1.5046769960859652
Initial weight: 1.0, Updated weight: 1.5580870819162018
Initial weight: 1.0, Updated weight: 0.554406040254968
Initial weight: 1.0, Updated weight: 0.5544062835543123
Initial weight: 1.0, Updated weight: 0.5555461935366892
Initial weight: 1.0, Updated weight: 0.632456315445355
Initial weight: 1.0, Updated weight: 1.2001792723059206
Initial weight: 1.0, Updated weight: 1.5398255566140917
Plot: Weight change as a function of frequency
[38]:
%matplotlib inline
fig, axes = plt.subplots()
ls1 = axes.plot(freqs, dw_dict_nn[delta_t[0]], color='r')
ls2 = axes.plot(freqs, dw_dict_nn[delta_t[1]], linestyle='--', color='r')
axes.plot(freqs, dw_dict[delta_t[0]], color='b')
axes.plot(freqs, dw_dict[delta_t[1]], linestyle='--', color='b')
axes.set_xlabel(r'${\rho}(Hz)$')
axes.set_ylabel(r'${\Delta}w$')
lines = axes.get_lines()
legend = plt.legend([lines[i] for i in [0,2]], ['Nearest spike', 'All-to-all'])
legend2 = plt.legend([ls1[0], ls2[0]], [r'${\Delta}t = 10ms$', r'${\Delta}t = -10ms$'], loc = 4)
axes.add_artist(legend)
[38]:
<matplotlib.legend.Legend at 0x7fec77d9bc70>
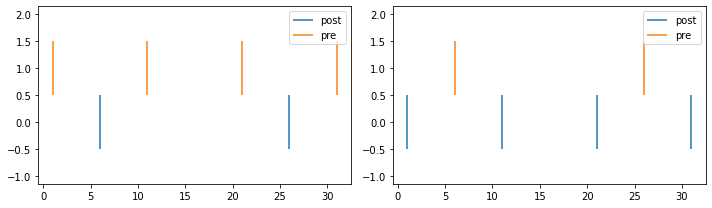
Triplet protocol
In the first triplet protocol, each triplet consists of two presynaptic spikes and one postsynaptic spike separated by \(\Delta{t_1} = t^{post} - t_1^{pre}\) and \(\Delta{t_2} = t^{post} - t_2^{pre}\) where \(t_1^{pre}\) and \(t_2^{pre}\) are the presynaptic spikes of the triplet.
The second triplet protocol is similar to the first with the difference that it contains two postsynaptic spikes and one presynaptic spike. In this case, \(\Delta{t_1} = t_1^{post} - t^{pre}\) and \(\Delta{t_2} = t_2^{post} - t^{pre}\).
[15]:
def create_triplet_spikes(_delays, n, trip_delay=1):
_dt1 = abs(_delays[0])
_dt2 = abs(_delays[1])
_interval = _dt1 + _dt2
# pre_spikes
start = 1
stop = 0
pre_spike_times = []
for i in range(n):
start = stop + 1 if i == 0 else stop + trip_delay
stop = start + _interval
pre_spike_times = np.hstack([pre_spike_times, [start, stop]])
# post_spikes
start = 1 + _dt1
step = _interval + trip_delay
stop = start + n * step
post_spike_times = np.arange(start, stop, step).astype(float)
return pre_spike_times, post_spike_times
[16]:
pre_spike_times, post_spike_times = create_triplet_spikes((5, -5), 2, 10)
l = []
l.append(post_spike_times)
l.append(pre_spike_times)
colors1 = ['C{}'.format(i) for i in range(2)]
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 3))
ax1.eventplot(l, colors=colors1)
ax1.legend(['post', 'pre'])
l = []
l.append(pre_spike_times)
l.append(post_spike_times)
ax2.eventplot(l, colors=colors1)
ax2.legend(['post', 'pre'])
plt.tight_layout()
STDP model with triplets
The standard STDP model fails to reproduce the triplet experiments shown in [2] and [3]. The experments show a clear asymmetry between the pre-post-pre and post-pre-post protocols, but the standard STDP rule fails to reproduce it.
Triplet model with triplet protocol
Let us try to formulate the two triplet protocols using the triplet model and check if we can reproduce the asymmetry in the change in weights.
[17]:
# Simulation
def run_triplet_protocol_simulation(neuron_model_name, synapse_model_name,
neuron_opts, nest_syn_opts,
spike_delays, n_triplets = 1, triplet_delay = 1000,
pre_post_pre=True):
dw_vec= []
# For pre-post-pre triplets
for _delays in spike_delays:
pre_spike_times, post_spike_times = create_triplet_spikes(_delays, n_triplets, triplet_delay)
if not pre_post_pre: # swap the spike arrays
post_spike_times, pre_spike_times = pre_spike_times, post_spike_times
sim_time = max(np.amax(pre_spike_times), np.amax(post_spike_times)) + 10. + 3 * syn_opts["delay"]
print('Simulating for (delta_t1, delta_t2) = ({}, {})'.format(_delays[0], _delays[1]))
dw = run_triplet_stdp_network(neuron_model_name, synapse_model_name,
neuron_opts, nest_syn_opts,
pre_spike_times=pre_spike_times,
post_spike_times=post_spike_times,
sim_time=sim_time)
dw_vec.append(dw)
return dw_vec
[18]:
# All to all - Parameters are taken from [4]
syn_opts = {
'delay': 1.,
'tau_minus': 33.7,
'tau_plus': 16.8,
'tau_x': 946.,
'tau_y': 27.,
'A2_plus': 6.1e-3,
'A3_plus': 6.7e-3,
'A2_minus': 1.6e-3,
'A3_minus': 1.4e-3,
'Wmax': 50.,
'Wmin' : 0.,
'w': 1.
}
synapse_suffix = neuron_model_name[neuron_model_name.find(nestml_synapse_model_name):]
neuron_opts_nn = {'tau_minus__for_' + synapse_suffix: syn_opts['tau_minus'],
'tau_y__for_' + synapse_suffix: syn_opts['tau_y']}
nest_syn_opts = syn_opts.copy()
nest_syn_opts.pop('tau_minus')
nest_syn_opts.pop('tau_y')
[18]:
27.0
[19]:
# Nearest spike - Parameters are taken from [4]
syn_opts_nn = {
'delay': 1.,
'tau_minus': 33.7,
'tau_plus': 16.8,
'tau_x': 575.,
'tau_y': 47.,
'A2_plus': 4.6e-3,
'A3_plus': 9.1e-3,
'A2_minus': 3e-3,
'A3_minus': 7.5e-9,
'Wmax': 50.,
'Wmin' : 0.,
'w': 1.
}
synapse_suffix_nn = neuron_model_name_nn[neuron_model_name_nn.find(nestml_synapse_model_name_nn):]
neuron_opts_nn = {'tau_minus__for_' + synapse_suffix_nn: syn_opts_nn['tau_minus'],
'tau_y__for_' + synapse_suffix_nn: syn_opts_nn['tau_y']}
nest_syn_opts_nn = syn_opts_nn.copy()
nest_syn_opts_nn.pop('tau_minus')
nest_syn_opts_nn.pop('tau_y')
[19]:
47.0
pre-post-pre triplet
[20]:
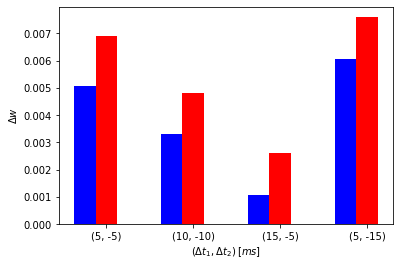
pre_post_pre_delays = [(5, -5), (10, -10), (15, -5), (5, -15)]
# All-to-All interation
dw_vec = run_triplet_protocol_simulation(neuron_model_name, synapse_model_name,
neuron_opts, nest_syn_opts,
pre_post_pre_delays,
n_triplets=1,
triplet_delay=1000)
# Nearest spike interaction
dw_vec_nn = run_triplet_protocol_simulation(neuron_model_name_nn, synapse_model_name_nn,
neuron_opts_nn, nest_syn_opts_nn,
pre_post_pre_delays,
n_triplets=1,
triplet_delay=1000)
Simulating for (delta_t1, delta_t2) = (5, -5)
Initial weight: 1.0, Updated weight: 1.0050613336422116
Simulating for (delta_t1, delta_t2) = (10, -10)
Initial weight: 1.0, Updated weight: 1.0033041134126772
Simulating for (delta_t1, delta_t2) = (15, -5)
Initial weight: 1.0, Updated weight: 1.0010569740202522
Simulating for (delta_t1, delta_t2) = (5, -15)
Initial weight: 1.0, Updated weight: 1.0060708925921593
Simulating for (delta_t1, delta_t2) = (5, -5)
Initial weight: 1.0, Updated weight: 1.0069212695313912
Simulating for (delta_t1, delta_t2) = (10, -10)
Initial weight: 1.0, Updated weight: 1.0048211690063567
Simulating for (delta_t1, delta_t2) = (15, -5)
Initial weight: 1.0, Updated weight: 1.0026215076729108
Simulating for (delta_t1, delta_t2) = (5, -15)
Initial weight: 1.0, Updated weight: 1.0076053401541742
[21]:
# Plot
bar_width = 0.25
fig, axes = plt.subplots()
br1 = np.arange(len(pre_post_pre_delays))
br2 = [x + bar_width for x in br1]
axes.bar(br1, dw_vec, width=bar_width, color='b')
axes.bar(br2, dw_vec_nn, width=bar_width, color='r')
axes.set_xticks([r + bar_width for r in range(len(br1))])
axes.set_xticklabels(pre_post_pre_delays)
axes.set_xlabel(r'${(\Delta{t_1}, \Delta{t_2})} \: [ms]$')
axes.set_ylabel(r'${\Delta}w$')
plt.show()
post-pre-post triplet
[22]:
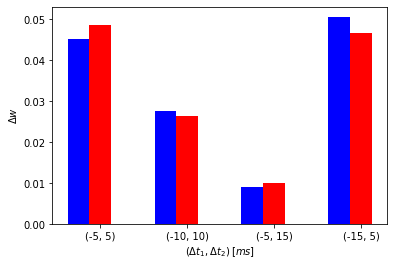
post_pre_post_delays = [(-5, 5), (-10, 10), (-5, 15), (-15, 5)]
# All-to-All interaction
dw_vec = run_triplet_protocol_simulation(neuron_model_name, synapse_model_name,
neuron_opts, nest_syn_opts,
post_pre_post_delays,
n_triplets=10,
triplet_delay=1000,
pre_post_pre=False)
# Nearest spike interaction
dw_vec_nn = run_triplet_protocol_simulation(neuron_model_name_nn, synapse_model_name_nn,
neuron_opts_nn, nest_syn_opts_nn,
post_pre_post_delays,
n_triplets=10,
triplet_delay=1000,
pre_post_pre=False)
Simulating for (delta_t1, delta_t2) = (-5, 5)
Initial weight: 1.0, Updated weight: 1.0452168105331474
Simulating for (delta_t1, delta_t2) = (-10, 10)
Initial weight: 1.0, Updated weight: 1.0275785817728278
Simulating for (delta_t1, delta_t2) = (-5, 15)
Initial weight: 1.0, Updated weight: 1.008936270857372
Simulating for (delta_t1, delta_t2) = (-15, 5)
Initial weight: 1.0, Updated weight: 1.050539844879153
Simulating for (delta_t1, delta_t2) = (-5, 5)
Initial weight: 1.0, Updated weight: 1.048644757755009
Simulating for (delta_t1, delta_t2) = (-10, 10)
Initial weight: 1.0, Updated weight: 1.026345906763637
Simulating for (delta_t1, delta_t2) = (-5, 15)
Initial weight: 1.0, Updated weight: 1.0099778920748412
Simulating for (delta_t1, delta_t2) = (-15, 5)
Initial weight: 1.0, Updated weight: 1.0466078732990223
[23]:
# Plot
bar_width = 0.25
fig, axes = plt.subplots()
br1 = np.arange(len(pre_post_pre_delays))
br2 = [x + bar_width for x in br1]
axes.bar(br1, dw_vec, width=bar_width, color='b')
axes.bar(br2, dw_vec_nn, width=bar_width, color='r')
axes.set_xticks([r + bar_width for r in range(len(br1))])
axes.set_xticklabels(post_pre_post_delays)
axes.set_xlabel(r'${(\Delta{t_1}, \Delta{t_2})} \: [ms]$')
axes.set_ylabel(r'${\Delta}w$')
plt.show()
Quadruplet protocol
This protocol is with a set of four spikes and defined as follows: a post-pre pair with a delay of \(\Delta{t_1} = t_1^{post} - t_1^{pre}< 0\) is followed after a time \(T\) with a pre-post pair with a delat of \(\Delta{t_2} = t_2^{post} - t_2^{pre} > 0\). When \(T\) is negative, a pre-post pair with a delay of \(\Delta{t_2} = t_2^{post} - t_2^{pre} > 0\) is followed by a post-pre pair with delay \(\Delta{t_1} = t_1^{post} - t_1^{pre}< 0\).
Try to formulate this protocol using the defined model and reproduce the following weight change window graph.
References
[1] Bi G, Poo M (1998) Synaptic modifications in cultured hippocampal neurons: dependence on spike timing, synaptic strength, and postsynaptic cell type. J Neurosci 18:10464–10472.
[2] Wang HX, Gerkin RC, Nauen DW, Bi GQ (2005) Coactivation and timing-dependent integration of synaptic potentiation and depression. Nat Neurosci 8:187–193.
[3] Sjöström P, Turrigiano G, Nelson S (2001) Rate, timing, and cooperativity jointly determine cortical synaptic plasticity. Neuron 32:1149–1164.CrossRefPubMedGoogle Scholar
[4] J.P. Pfister, W. Gerstner Triplets of spikes in a model of spike-timing-dependent plasticity J. Neurosci., 26 (2006), pp. 9673-9682
Acknowledgements
This software was developed in part or in whole in the Human Brain Project, funded from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under Specific Grant Agreements No. 720270 and No. 785907 (Human Brain Project SGA1 and SGA2).
License
This notebook (and associated files) is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 2 of the License, or (at your option) any later version.
This notebook (and associated files) is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
[ ]: